Technology
The Serum Epitope Repertoire Analysis (SERA) platform is a universal serology platform that utilizes bacterial display peptide library technology and next generation sequencing to broadly profile antibody repertoires and identify the antigens and epitopes associated with many diseases – all in a single assay.
Benefits
Unlimited Multiplexing
Simultaneous profiling of multiple pathogens from a single specimen
Exquisitely High-Resolution
Epitope mapping in infectious diseases, oncology and autoimmune disease
Clinically Relevant Analysis
State-of-the-Art Database with 26,000+ profiled antibody repertoires from individuals with disease as well as healthy blood donors
Scientific Support
Leading experts in epitope analysis and bioinformatics
Ideal for
Epitope Mapping
Target Identification
Disease Stratification
Response Measurement
Serum Epitope Repertoire Analysis (SERA) Platform
Combining universal chemistry with proprietary informatics, SERA assesses antibody repertoires to address research and clinical needs.
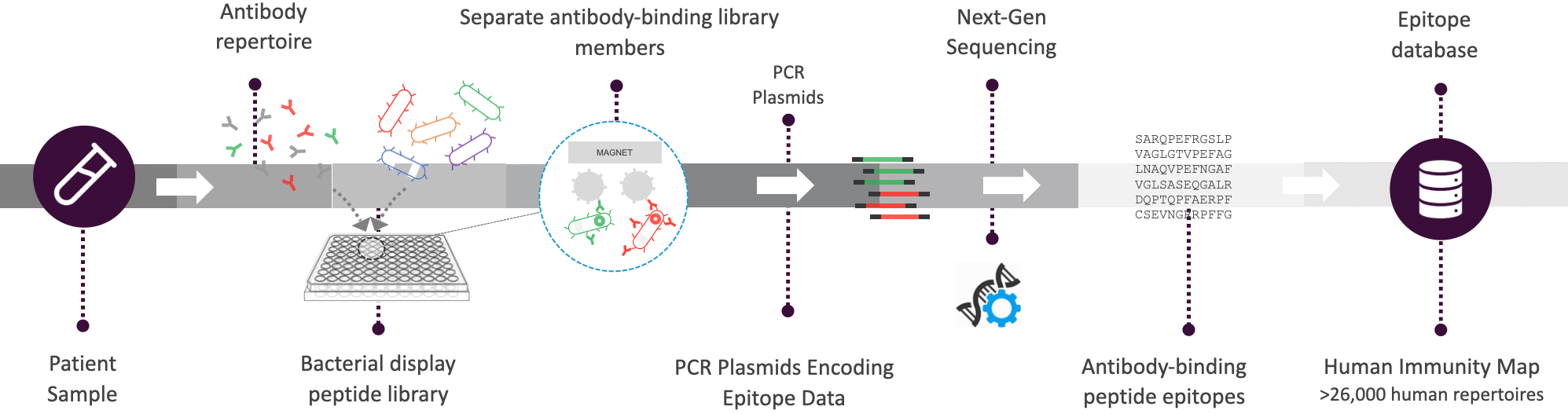
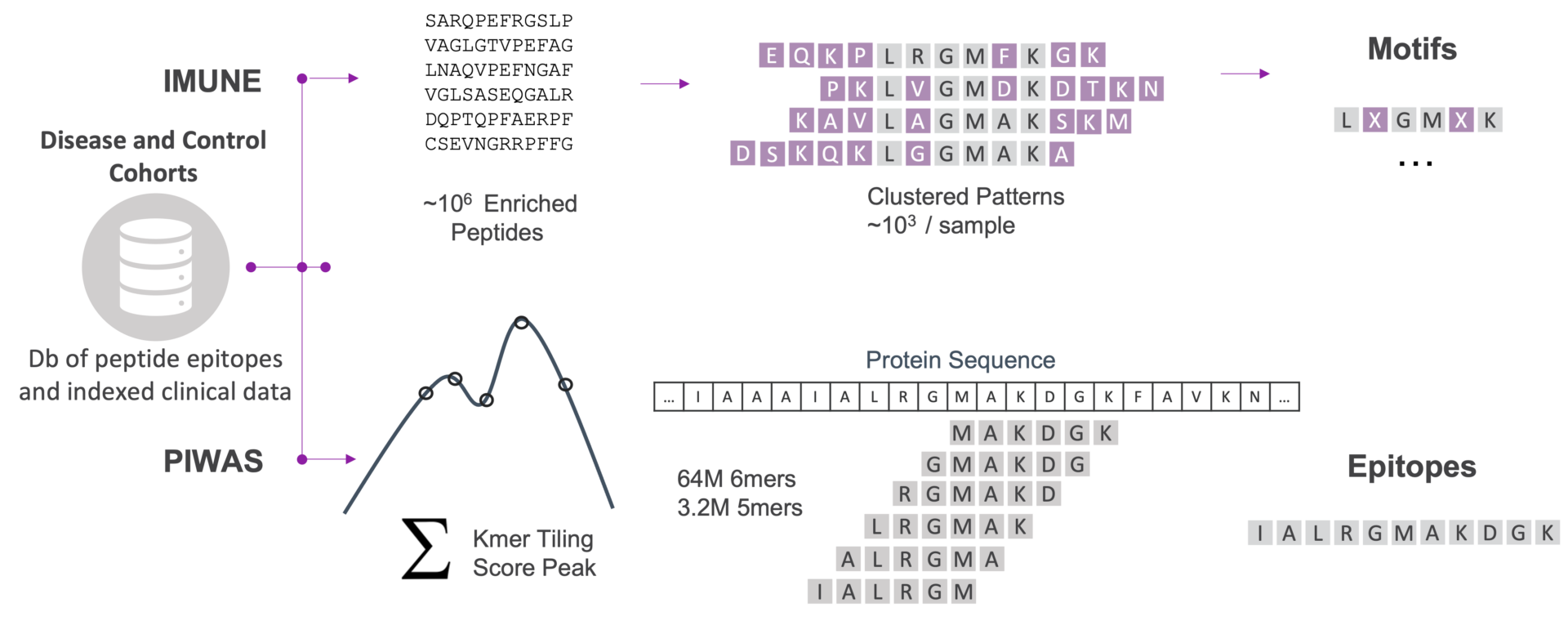
Two Complementary Bioinformatics Methods
Identify epitopes (PIWAS) and motifs (IMUNE) associated with disease.
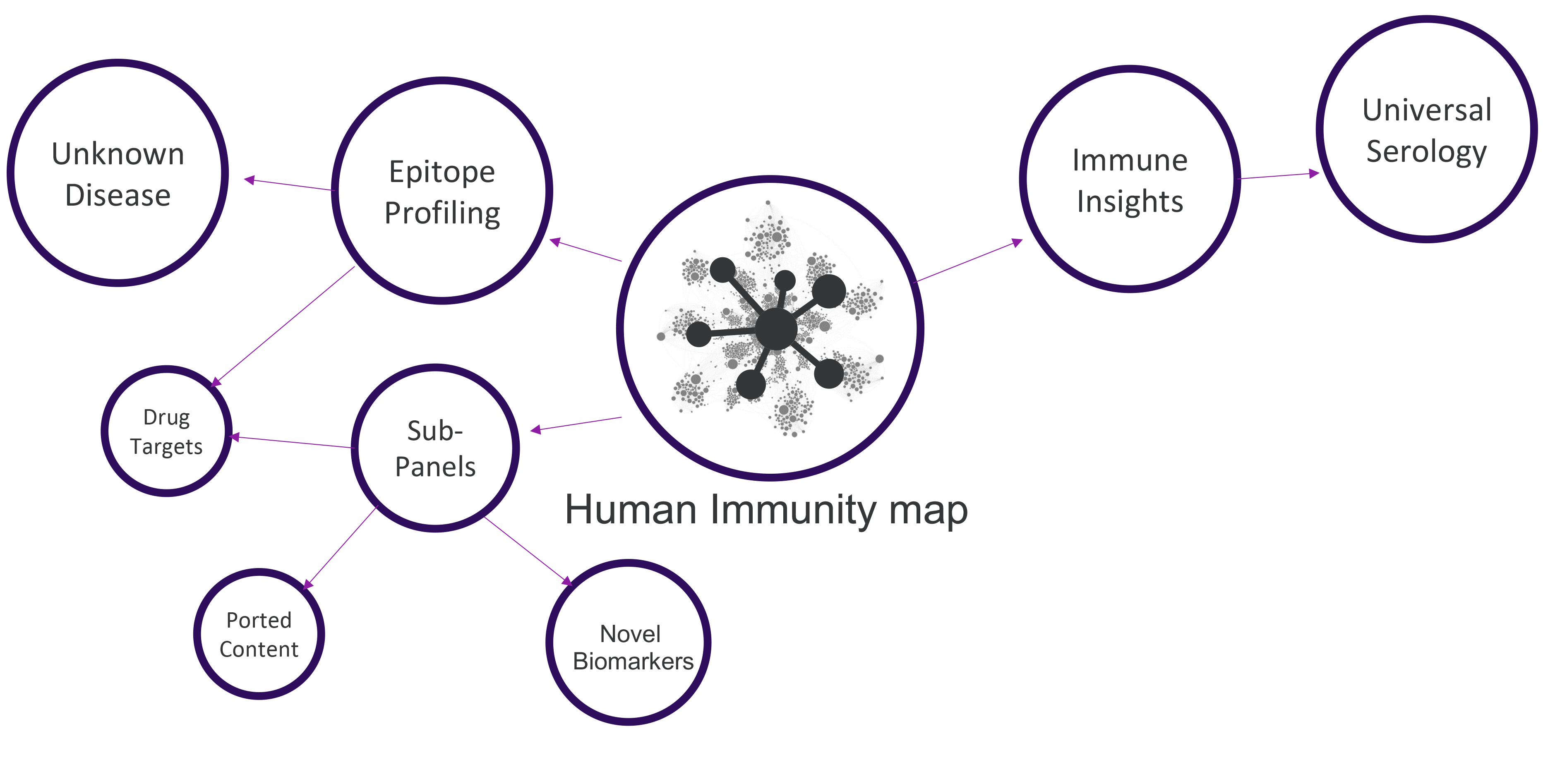
Applications
SERA provides value across multiple areas of Academic, Industry and Government research.
Disease Categories
Autoimmune Disease
Oncology
Infectious Disease
Outcomes
Uncover novel epitopes
Discover unique and specific antigens
Unmask disease associations
Develop novel biomarkers
Profile patients' immune response
Stratify patients
Partnering
Serimmune partners with academic institutions, biopharma and healthcare organizations and government that seek to power immune-related research, product development and to make healthcare decisions.
The SERA platform has features that make it robust and ideal for biomarker discovery, clinical diagnostic panel development, serologic surveillance.
Autoimmune Disease
SERA has been used to identify both validated and potentially novel antigens in multiple autoimmune disorders. Candidate autoantigens in cohorts may help distinguish sub-types of disease or help to predict/track response to therapy, identify new targets, and enable precision immunology.
Oncology
Using SERA, researchers can identify candidate autoantigens that may be used as biomarkers for diagnosis, sub-typing, or monitoring in oncology. B cell response in immuno-oncology has recently been highlighted in association with response to checkpoint inhibitor therapy. SERA enables the tracking of potential autoantigen signals before and after therapy to identify candidate biomarkers of response to therapy, paving the way for precision oncology.
Infectious Disease
SERA can identify the most significant antigens in infections for diagnostic and therapeutic target discovery as demonstrated in COVID-19. Using cohorts with disease, SERA can also identify sensitive and specific peptides for infections that can be used on alternate diagnostic platforms.
Therapeutics
SERA is a powerful tool that pharmaceutical and biotech organizations can leverage for identification of motifs and peptide epitopes recognized by monoclonal antibodies. These can be used to optimize the selection of therapeutic candidates. SERA has also identified the antigenic targets of antibody repertoires to identify therapeutic targets. This screening may identify potential off-target antigens for our partners.
Population Serosurveillance
SERA has the broad potential to enable dynamic monitoring and surveillance for hundreds of diseases in one assay. It has recently been used to demonstrate the increase in seropositivity early in the COVID-19 epidemic. Significantly, SERA has also demonstrated the ability to detect seropositivity retrospectively and in silico, without the need of any new assays, enabling serosurveillance in individuals and populations for a variety of emerging infections.
